NK cell - Dress Code for Killing
- Martin Döhring

- 28. Sept. 2025
- 2 Min. Lesezeit
...molecular choreography when a natural killer (NK) cell “gets dressed to kill” a tumor cell.
Molecular Steps of NK Cell–Mediated Tumor Cell Killing
1. Tumor Cell Recognition
NK cells don’t rely on a single receptor like TCR, but integrate signals from a complex set of activating and inhibitory receptors:
Inhibitory receptors (checkpoints for tolerance):
KIRs (Killer-cell immunoglobulin-like receptors) bind MHC-I (HLA-A/B/C).
NKG2A/CD94 binds HLA-E.→ If normal MHC-I is present, these receptors deliver ITIM signals → recruit SHP-1/2 phosphatases → block activation.
Activating receptors (danger sensors):
NKG2D recognizes MICA/MICB, ULBPs (stress-induced ligands).
DNAM-1 (CD226) recognizes CD112 (Nectin-2) and CD155 (PVR).
Natural cytotoxicity receptors (NCRs: NKp30, NKp44, NKp46) bind viral/tumor ligands.
CD16 (FcγRIIIa) mediates ADCC by binding IgG-coated tumor cells.
Balance of signals decides: If inhibitory input is low (e.g., tumor cells often downregulate MHC-I) and activating input is high, the NK cell commits to kill.
2. Immunological Synapse Formation
Once activated, NK cells polarize toward the tumor cell:
Adhesion molecules:
LFA-1 (CD11a/CD18) on NK binds ICAM-1 on tumor.
Forms a stable immune synapse.
Inside NK cell:
Actin cytoskeleton reorganizes.
Microtubule organizing center (MTOC) reorients toward the synapse.
Lytic granules (containing perforin & granzymes) traffic to the contact site.
3. Cytotoxic Arsenal Release
NK cells have two main molecular killing strategies:
(A) Granule Exocytosis Pathway
Perforin inserts into tumor cell membrane → polymerizes → forms pores.
Granzymes (mainly Granzyme B, A, M) enter cytosol via perforin pores or endocytosis.
Granzyme B cleaves caspases (e.g., caspase-3, caspase-7) and BID → apoptosis.
Granzyme A triggers caspase-independent apoptosis via DNA damage (nuclease activation).
Serglycin stabilizes perforin/granzymes in lytic granules.
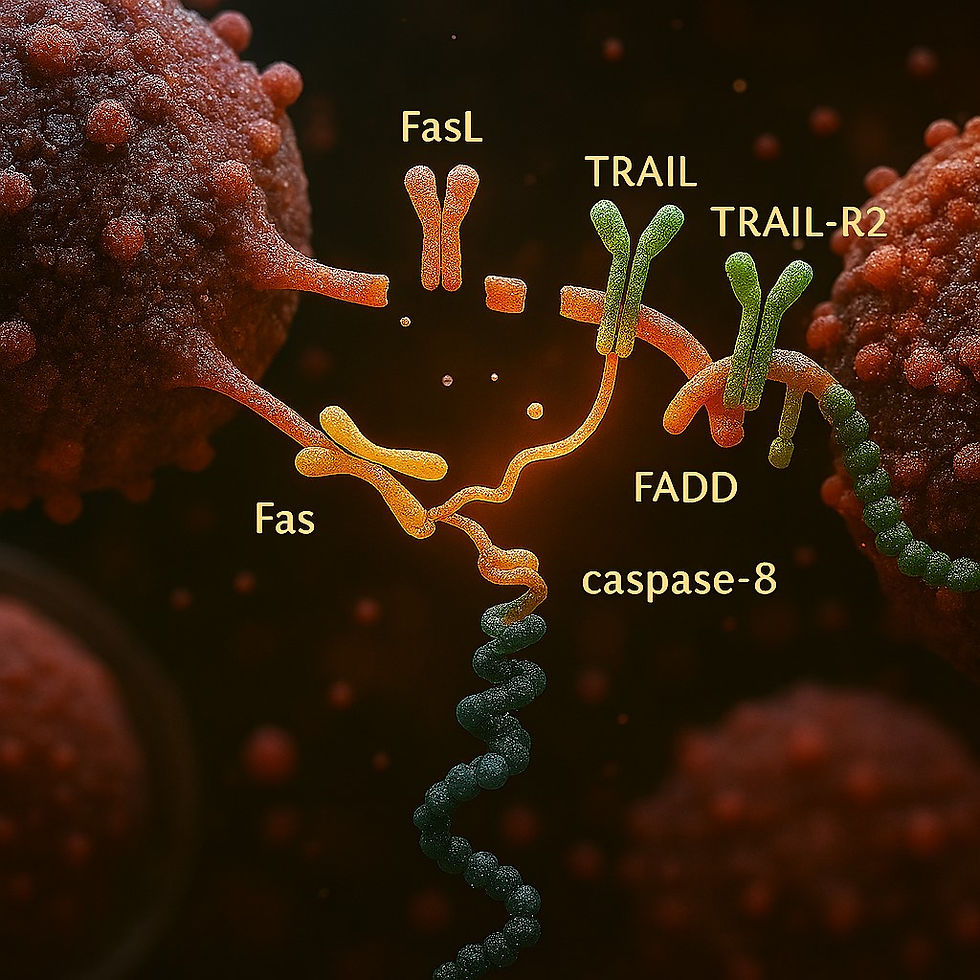
(B) Death Receptor Pathway
NK cells express FasL (CD95L) and TRAIL.
Bind Fas (CD95) and TRAIL-R1/R2 on tumor → recruit FADD → activate caspase-8 → apoptosis.
4. Cytokine and Chemokine Assault
NK cells also modulate the tumor microenvironment:
IFN-γ: Activates macrophages, boosts antigen presentation, shapes adaptive immunity.
TNF-α: Directly cytotoxic, pro-apoptotic via TNFR1.
GM-CSF, CCL3, CCL4, CCL5: Recruit dendritic cells, T cells, more NK cells.
5. Molecular Checkpoints & Tumor Evasion
Tumors evolve to resist NK attack:
Upregulating MHC-I → strengthens inhibitory KIR signals.
Shedding NKG2D ligands (MICA/B) → soluble decoys bind NKG2D and block activation.
Expressing PD-L1 → inhibits NK and T cells via PD-1.
Immunosuppressive cytokines (TGF-β, IL-10) → reduce NK receptor expression and cytotoxicity.
6. NK Cell “Dress Code for Killing”
When NK cells activate fully, their molecular “armor” includes:
Upregulated perforin + granzyme release machinery.
Polarized MTOC + lytic granules aligned to the synapse.
Enhanced cytotoxic receptors (NKG2D, NCRs, CD16).
Secretion of IFN-γ + TNF-α to orchestrate the immune assault.
✅ In summary:NK cells kill tumor cells by sensing missing-self (low MHC-I) and induced-self ligands (stress markers), forming a tight synapse, delivering perforin/granzymes or death ligands, and reinforcing with cytokine-mediated immune pressure. Tumor cells, in turn, deploy molecular tricks to escape this immune surveillance.







...let’s decode the molecular “dress code” Natural Killer (NK) cells wear when they’re ready to kill tumor cells. Think of it as their biochemical armor and weaponry, tailored for precision strikes against abnormal cells.
Molecular Dress Code of NK Cells for Tumor Killing
NK cells don’t rely on a single receptor like T cells do. Instead, they integrate signals from a dynamic balance of activating and inhibitory receptors to decide whether to kill a target.
1. Tumor Cell Recognition: The Permission Slip
NK cells scan for abnormalities in surface molecules:
Inhibitory Receptors (Tolerance Checkpoints):
KIRs (Killer-cell Immunoglobulin-like Receptors) bind MHC-I (HLA-A/B/C).
NKG2A/CD94 binds HLA-E.
If normal MHC-I is present → “Don’t kill” signal.
Activating Receptors (Kill Signals):
NKG2D binds stress-induced ligands like MICA/MICB, ULBP1–6.
...the whole molecular NK–tumor battle: Let me now expand this into a detailed, stepwise molecular storyline, so you can see every switch, signal, and effector at work.
Molecular Steps of NK Cell–Mediated Tumor Cell Killing
1. Tumor Cell Recognition — Balancing “Kill” vs “Don’t Kill”
NK cells function on the “missing self + induced self” principle:
Inhibitory receptors = “brakes”
KIRs (Killer Immunoglobulin-like Receptors) → bind classical MHC-I (HLA-A/B/C).
NKG2A/CD94 → binds HLA-E (non-classical MHC-I).
Signal: Engagement → ITIM (Immunoreceptor Tyrosine-based Inhibitory Motif) phosphorylation → recruits SHP-1/2 phosphatases → dephosphorylate Vav1, SLP-76, LAT → block cytotoxic cascades.
Activating receptors = “accelerators”
NKG2D → recognizes stress-induced ligands (MICA, MICB, ULBPs).
DNAM-1 (CD226) → binds CD112 (Nectin-2) and CD155 (PVR).
NCRs: NKp30,…